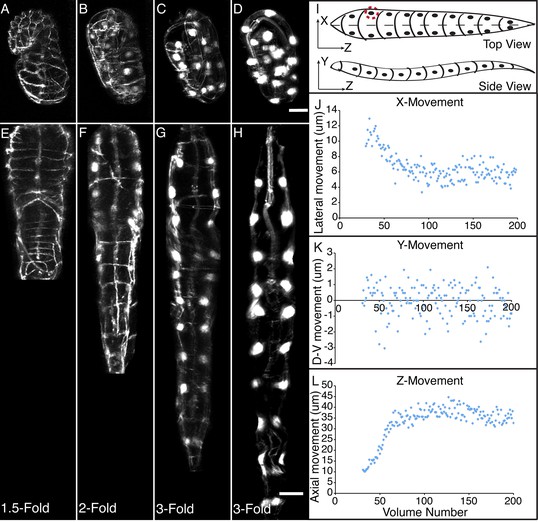
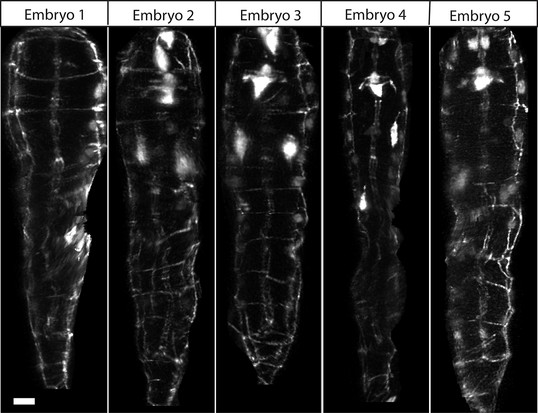
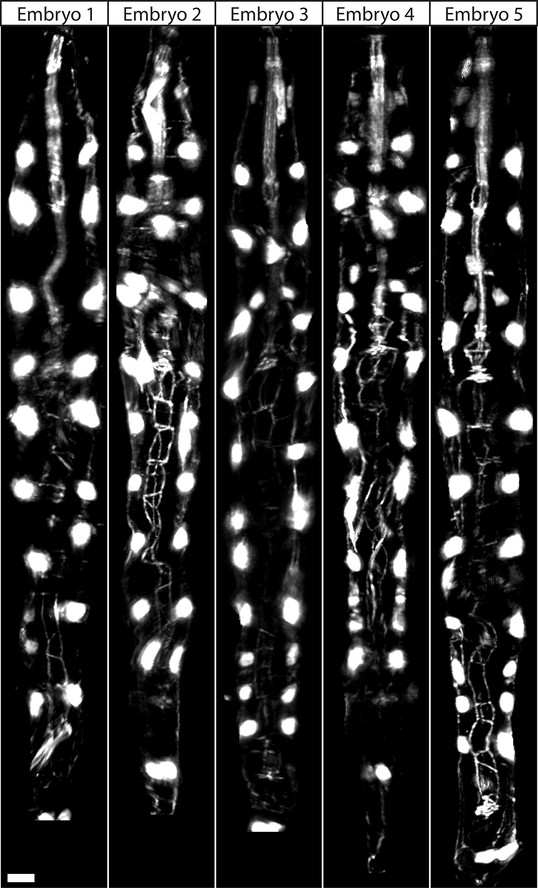
WORM UNTWISTING THE CAENORHABDITIS ELEGANS EMBRYO
PROJECT COLLABORATORS
Section on High Resolution Optical Imaging, NIBIB:
Hari Shroff, Ph.D, Ryan Patrick Christensen, Ph.D, Yicong Wu, Ph.D, Min Guo, Ph.D, Abhishek Kumar, Ph.D, Peter W Winter, Ph.D, Nicole Tashakkori.
Developmental Biology Program, Sloan-Kettering Institute:
Anthony Santella, Zhirong Bao.
Program in Cellular Neuroscience, Neurodegeneration and Repair, Department of Cell Biology, Yale University School of Medicine:
Javier Marquina-Solis, Ismar Kovacevic, Daniel A Colón-Ramos.
State Key Laboratory of Modern Optical Instrumentation, College of Optical Science and Engineering, Zhejiang University:
Huafeng Liu.
Department of Genetics and Developmental Biology, University of Connecticut Health Center:
William Mohler, Ph.D.