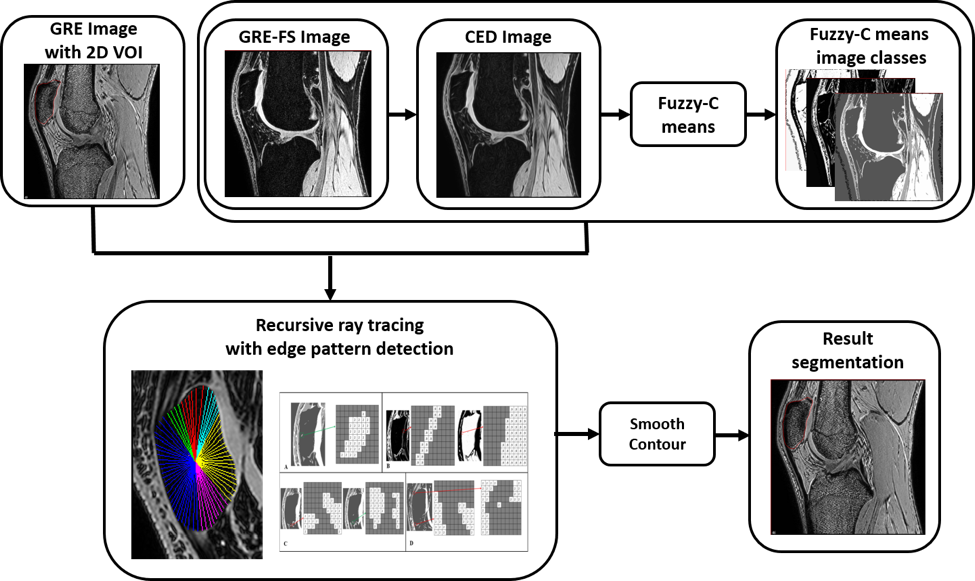
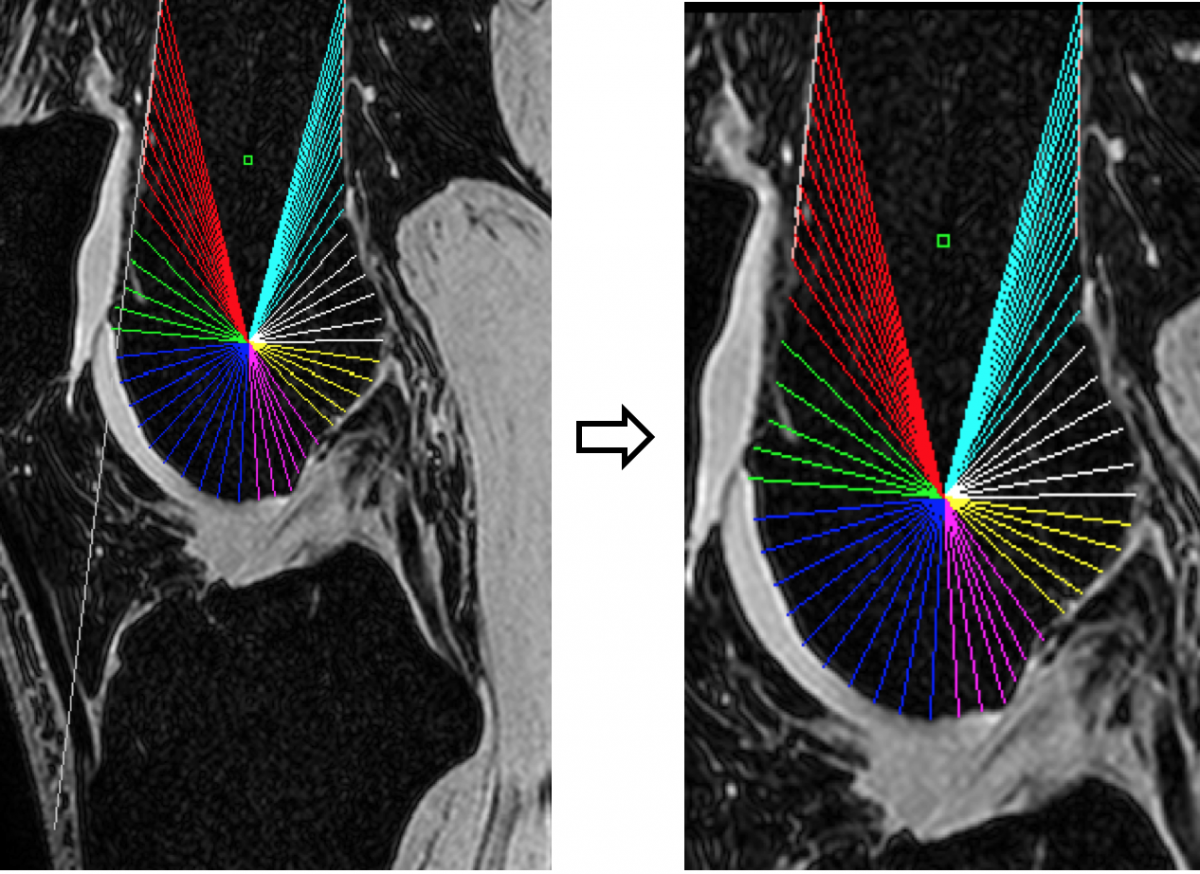
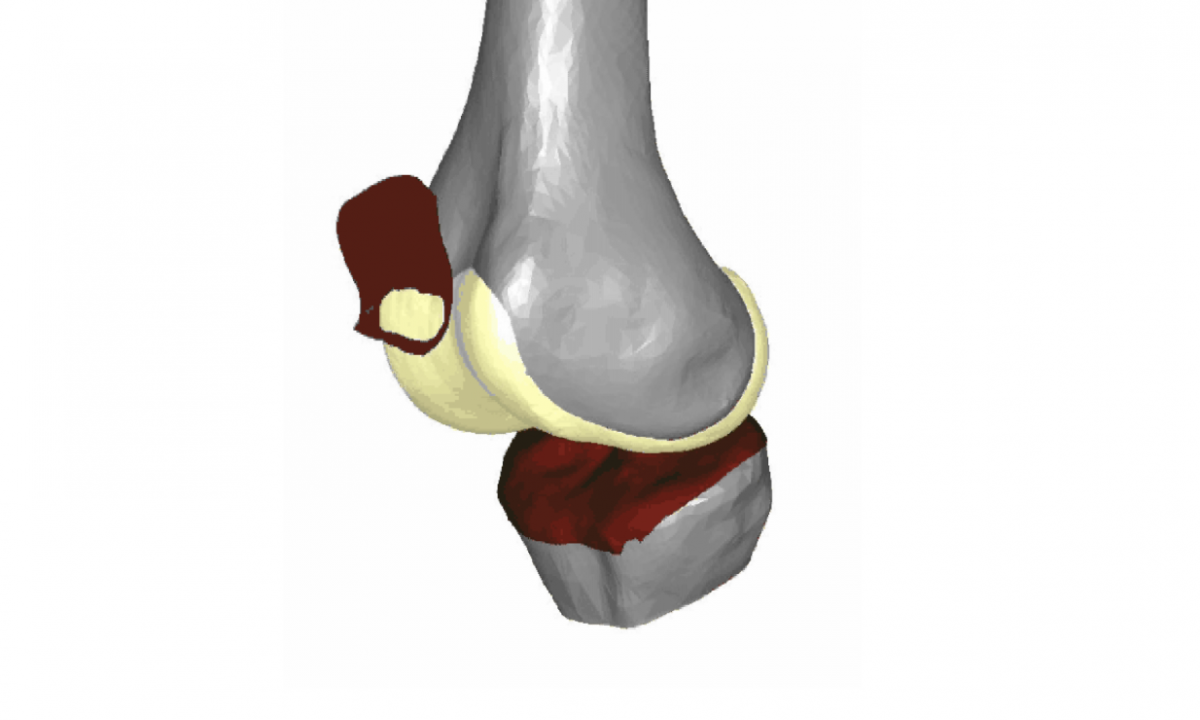
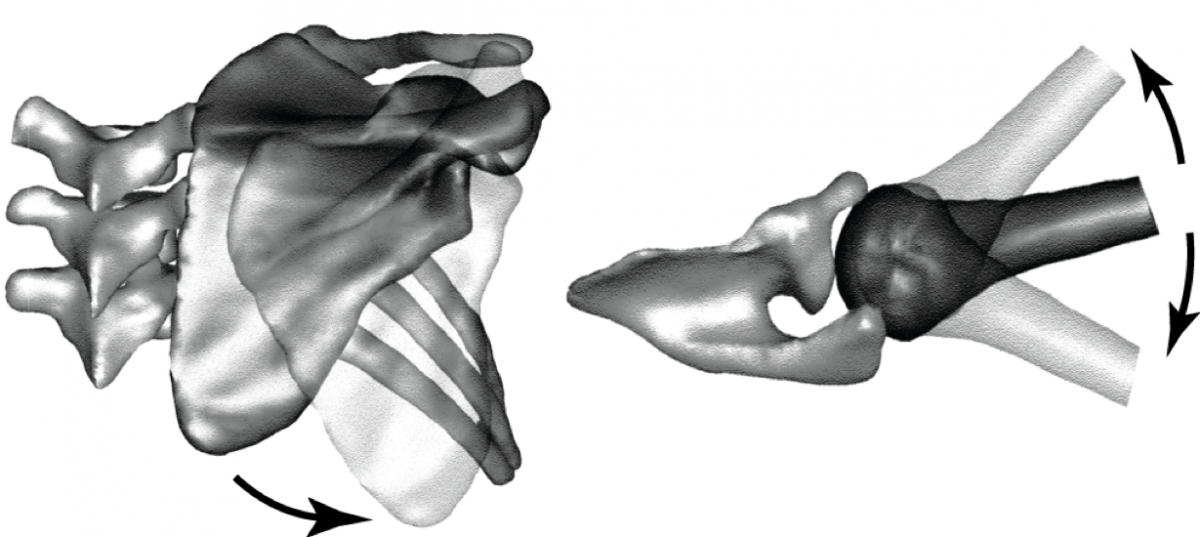
MRI KNEES SEGMENTATION
PROJECT COLLABORATORS
National Institutes of Neurological Disorders and Stroke:
Frances T(Sheehan) Gavelli, PhD (lab chief), Jennifer Jackson, PhD.
Previous:
NIH, Clinical Center, Functional and Applied Biomechanics Section, Rehabilitation Medicine Department.